Pathology, a branch of medicine focused on studying and diagnosing diseases, has witnessed significant advancements in recent decades, particularly with the integration of digital imaging technology. The introduction of digital cameras in microscopes has revolutionized the field, allowing pathologists to capture and analyze high-resolution images of organs, tissues, and fluids. This digitalization of histological images, initially in the form of photographs or videos, has evolved into a more sophisticated approach known as Whole Slide Imaging (WSI) in Digital Pathology (DP).
WSI technology emerged as a result of efforts to digitize entire tissue slides into highly detailed images. The concept of a “virtual microscope” was first introduced by Ferreira et al. in 1997, but it wasn’t until 1999 that the cost and speed of WSI systems became affordable. This breakthrough was made possible by the contributions of Wetzel and Gilbertson while working for Interscope Technologies. Since then, there has been a growing interest in the commercial use of WSI systems.
A WSI system typically consists of four main components: a light source, a microscope equipped with multiple lenses, a digital camera, and a mechanism for repositioning the camera view along the sample. These systems are capable of capturing high-resolution images in the gigapixel range, with the most commonly used magnifications being x20 or x40. In specific cases, higher magnifications are employed, such as for blood smears. WSI scanners can operate in different imaging modes, including bright-field, fluorescence, and multispectral imaging. Each mode highlights different anatomical structures or physiological events within the tissue, utilizing distinct light sources.
How Visualize WSIs?
To handle the vast amount of data generated by WSIs, a pyramid organization method is employed. Instead of storing the entire image in a single frame, the image is replicated at multiple resolutions, known as pyramid levels. Each level is then divided into two-dimensional blocks called tiles. Only the tiles corresponding to the viewing area are loaded into memory, optimizing the display of the high-resolution image. This approach overcomes the limitations of traditional single-frame organization, especially when dealing with extremely large images.
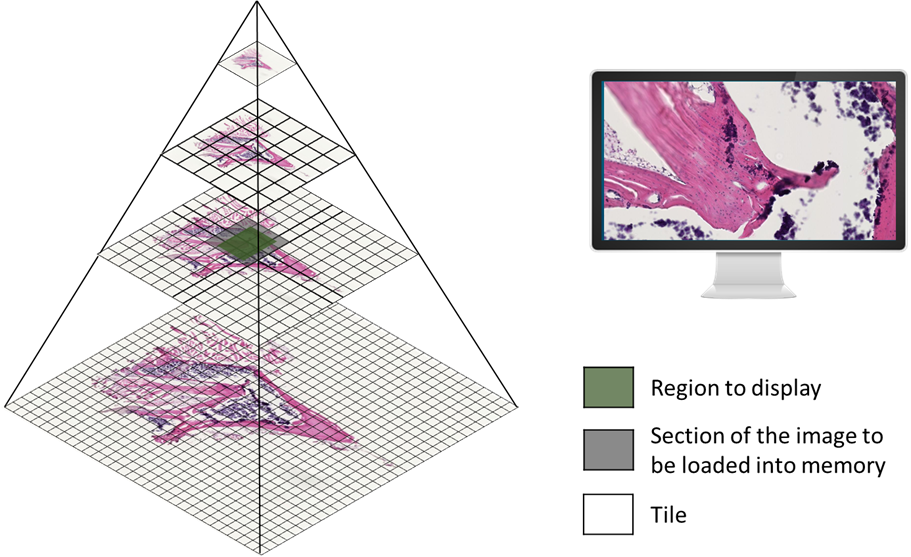
Figure 1: Pyramid organization used to store and visualize WSIs
Challenges Reading WSI files formats
Standardization of data formats for WSIs has been a challenge, leading to the development of several proprietary formats, such as NDPI, SVS, and SCN. However, two candidate formats, DICOM and TIFF, aim to become the industry standards. The DICOM format, developed by the DICOM Standard Committee Working Group 26, provides not only an image format but also protocols for image exchange and maintenance. Its wide adoption is expected due to the existing use of DICOM in radiology. Another contender is the TIFF format, particularly the variant created by the Open Microscopy Environment (OME) Consortium, which simplifies the use of single and multiple pyramid images by incorporating metadata.
Despite the advantages offered by the DICOM and TIFF formats, the transition from proprietary formats to the standard formats will be gradual due to the high costs associated with upgrading existing systems. To facilitate this transition, conversion tools, such as the Orthanc software, have been developed to convert WSIs to DICOM following the standardization guidelines. Not all platforms for viewing and analyzing digital pathological images perform this conversion, opting instead for libraries that enable the reading and writing of multiple formats. Two commonly used libraries are Bio-Formats, developed by the Open Microscopy Environment consortium, and OpenSlide, developed by Carnegie Mellon University. These libraries support the reading of various formats and are often used in combination to accommodate the different requirements of scanners.
Image Analysis Challenges
In addition to overcoming technical and format-related challenges, image analysis plays a crucial role in digital pathology. WSIs provide a wealth of data that can be extracted and analyzed at different levels, including cellular information and Tissue-based analysis. Cellular analysis (Object-level) involves classifying and categorizing different cell components, such as nuclei or cytoplasm, and identifying cell types or abnormalities within the tissue. Tissue-level analysis focuses on identifying and characterizing specific regions of interest within the tissue, such as necrotic areas or pre-neoplastic epithelium. Features can be extracted at various magnification levels, ranging from the pixel level to the object level and the tissue level, allowing for comprehensive analysis of tissue structures.
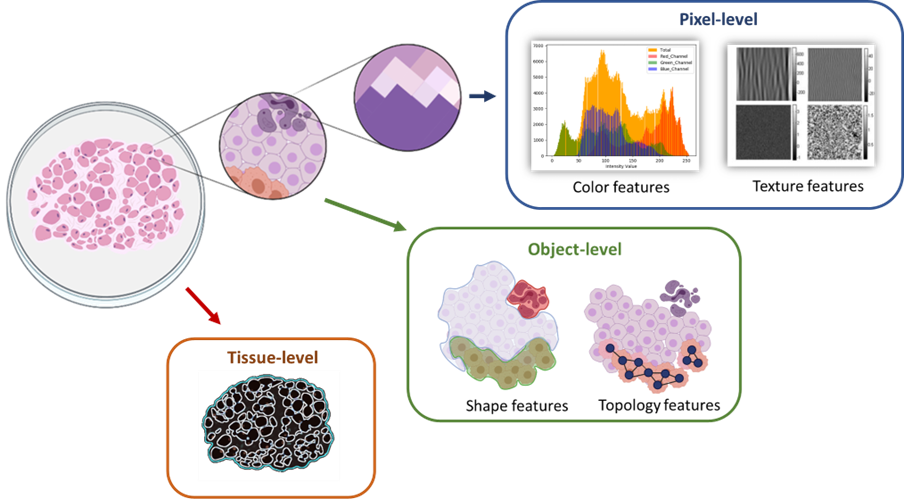
Figure 2: Categorizing of image analysis levels according to magnification.
However, image analysis in DP presents several challenges. The primary hurdle is the significant sample variation, which can be attributed to biological, pathological, and technological factors. Biological variation encompasses the diverse range of cells and tissue elements present in a sample, making accurate analysis and classification complex. Pathological variation arises from the different characteristics that tissue can exhibit due to inflammation, infection, or carcinogenesis. Technological variation stems from inconsistencies in image acquisition, staining techniques, and the expertise of technicians. These variations contribute to the complexity of image analysis and necessitate robust algorithms and techniques.
Another challenge in DP is the size of WSI files. With dimensions reaching the gigapixel scale, storing, transferring, and processing these files can be computationally demanding. To mitigate this challenge, modern analysis tools often focus on analyzing specific regions of interest rather than the entire slide, optimizing computational resources.
To address the challenges in image analysis, machine learning approaches have gained prominence. Supervised deep learning algorithms, in particular, have demonstrated remarkable results in analyzing, segmenting, and classifying microscopic tissue features. However, the success of these algorithms heavily relies on the availability of annotated data, also known as ground truth. Annotating pathological images requires expert knowledge in the specific disease area, posing difficulties in acquiring comprehensive datasets. Moreover, datasets frequently contain sensitive clinical information, limiting their availability for research purposes. Consequently, some analysis tools for WSIs do not currently incorporate supervised algorithms.
Conclusion
Digital pathology, driven by the integration of digital imaging technology, has transformed the field of pathology. Whole Slide Imaging (WSI) has enabled pathologists to capture, digitize, and analyze high-resolution images of tissue samples. However, challenges such as sample variation, large file sizes, and the need for annotated data persist. Machine learning approaches, especially supervised deep learning algorithms, show promise in overcoming these challenges. Nonetheless, the scarcity of annotated datasets and the complexity of pathological image analysis present ongoing hurdles. Digital pathology continues to evolve, offering new opportunities for disease diagnosis, research, and improved patient care.
BMD in DP image analysis
In the realm of addressing these challenges, BMD Software has been at the forefront, focusing on the enhancement of archive systems that adhere to the DICOM standard. Through our dedicated efforts, we have developed software solutions that enable the seamless visualization and navigation of histological images with exceptional levels of detail and resolution. These advancements empower pathologists to analyze and interpret digital slides in a collaborative environment, facilitating interdisciplinary collaborations and second opinions.
Furthermore, we have actively engaged in research and development endeavors, with a specific focus on the development of cutting-edge algorithms for nucleus detection and classification.
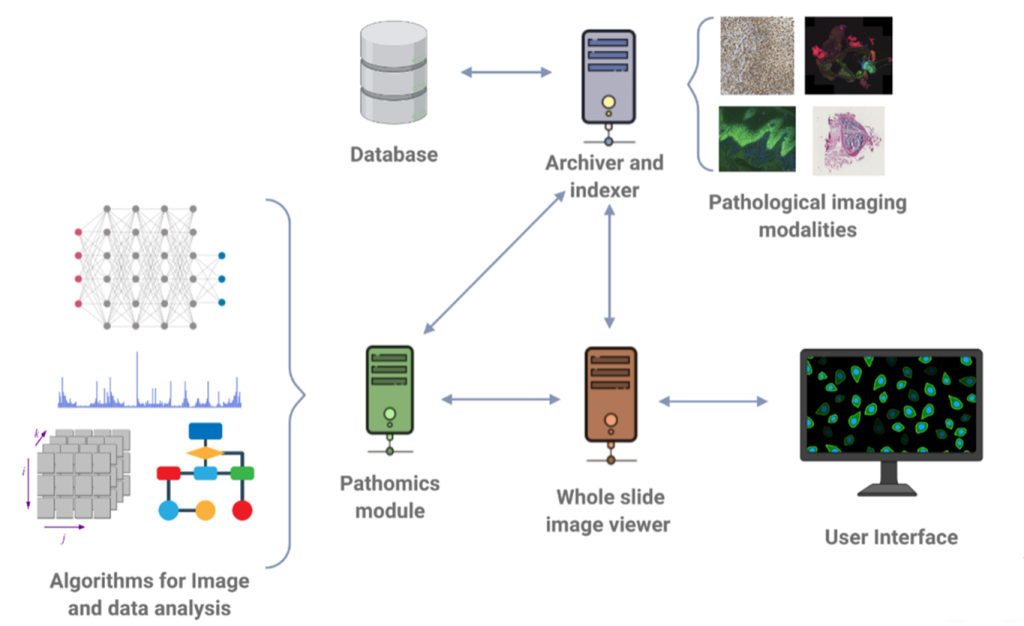
Figure 3: Research and development focused on Digital Pathology.
At BMD Software, we recognize the critical role that digital pathology plays in revolutionizing disease diagnosis and treatment. By addressing the challenges associated with sample variation, large file sizes, and the scarcity of annotated data, we aim to empower pathologists with state-of-the-art tools and technologies that streamline their workflow, improve diagnostic accuracy, and ultimately enhance patient outcomes.
