Savvy provides applications and services for highly-accurate and assisted biomedical information extraction from scientific literature, patents and electronic health records.
Why Savvy?
Highly accurate
Automatic extraction algorithms are heavily evaluated and compared with similar solutions, always achieving high performance results.
Easy to use
Graphical user interfaces, REST services and CLI tools are straightforward and self-explanatory to use, also accompanied with detailed documentation.
Customizable
Knowledge-base resources and complex processing pipeline steps can be optimized considering specific goals, focusing on target biomedical sub-domains.
Award-winning technology.
Savvy components participated in international challenges targeting biomedical information extraction tasks, always achieving the top results.
Named entity recognition
Named entity recognition components always presented the best results on the market, when compared against both research and commercial tools. The results achieved in multiple corpora and challenges reflect this fact.
Assisted curation
The innovative user interfaces presented a breakthrough on the field, with simple and self-explanatory interactions. The results achieved on the BioCreative challenges reflect this fact.
Fast and accurate
High accuracy and fast processing times, provide state-of-the-art automatic extraction and indexing of biomedical entities and their relations from literature, patents and electronic health records.
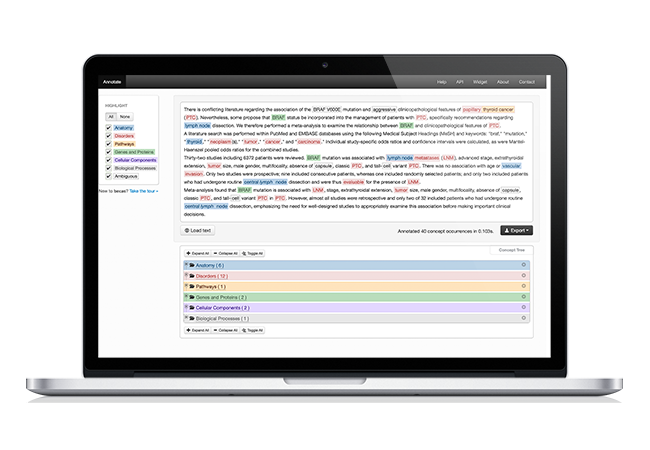
Biomedical concept recognition
Concept recognition is provided through dictionary matching and machine learning with normalization methods, supporting automatic extraction of species, cell, cellular components, gene and proteins, chemicals, biological processes, molecular functions, disorders, and anatomical entities. The most popular input and output formats are supported, namely raw text, PDF, Pubmed XML, BioC, CoNLL, A1 and JSON.
Concept recognition features are available through a Web Page for simplified and interactive usage, REST services for programming access, and CLI tool for rapid annotation.
Biomedical relation extraction
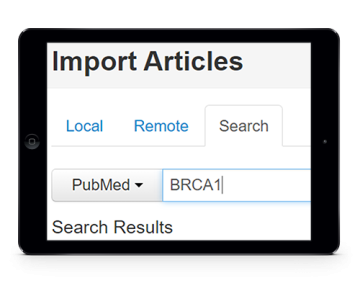
After collecting concept names and respective identifiers from literature, one may also collect the relations between such concepts that are referred in the documents. For instance, protein-protein interactions and relations between disorders and drugs can be automatically extracted from texts.
Relation extraction services can be used from the assisted curation tool or through REST services.

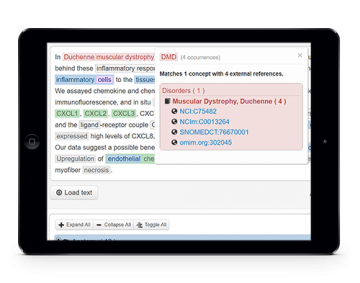
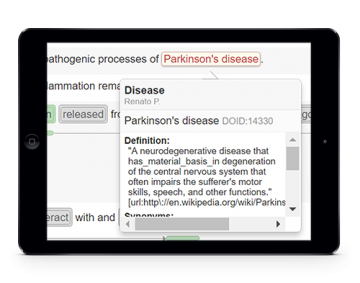
Assisted document curation
Biomedical text mining and assisted curation is available through a web-based platform with highly usable interfaces for manual and automatic in-line annotation of concepts and relations. A comprehensive set of de facto standard knowledge bases are integrated and indexed to provide straightforward concept normalization features. Moreover, real-time collaboration and conversation functionalities allow discussing details of the annotation task as well as providing instant feedback of curators interactions. In the end, in order to collect the curated annotations, features are available to export the resulting annotations using standard file formats.
When evaluated by expert curators in international challenges, the solution obtained highly positive results in terms of usability, reliability and performance.
Semantic search and indexing
When both concept names and relations are collected from documents, indexing can be applied to enable semantic search, allowing users to easily find the documents most relevant for their research. For instance, documents that present the relation between two specific concepts can be easily and rapidly accessible.
Multiple installation options available.
Cloud
Dramatically reduce IT costs by safely and securely outsourcing biomedical information extraction services.
Local servers
Install biomedical information extraction applications and services on local servers, enabling rapid extraction and indexing.


